- Home
- Kinase Assay Support Portal
- Kinase Assay Support Information
- Capillary Electrophoresis Assay Support
Capillary Electrophoresis Assay Support
Mobility Shift Assay (MSA) has been widely used in drug discovery and research for over 20 years as a method for separating substrates and products in enzymatic reactions. At Carna, we have optimized various elements involved in substrate-product separation and analysis—such as buffer conditions and separation parameters—to enable the use of SCIEX’s PA800 system for MSA. These optimized capillary electrophoresis assay conditions are applied in the QC testing of our in-house produced proteins.
Mobility Shift Assay (MSA) is an assay method that separates and detects substrates and products in enzymatic reactions based on differences in their mobility. When enzymes such as kinases act on a substrate, phosphorylation alters the charge of the product, resulting in a charge difference between the substrate and product. This difference leads to a variation in migration speed during electrophoresis, allowing detection of kinase phosphorylation activity.
Carna provides optimized assay conditions for Mobility Shift Assay (MSA)/Capillary Electrophoresis Kinase Assay using SCIEX’s PA800 system as the detection platform.
For information on the kinases corresponding to each FL-labeled Peptide Substrate listed in the table below, please refer to FL-labeled Peptide Substrates List page, or feel free to contact us from here or at info@sb.carnabio.com for further assistance.
| Experimental/lab equipment | Manufacturer | Product number |
|---|---|---|
| PA 800 Plus | SCIEX | A66528 |
| Bare Fused-Silica Capillary | SCIEX | 338451 |
| Polypropylene 96 well microplate | Greiner | 655201 |
| Substrate Name (FL-labeled Peptide Substrate) |
Substrate Cat. No. |
Voltage (KV) | Duration (min) | Pressure (psi) | Pressure direction | Peak Charge Figure |
|---|---|---|---|---|---|---|
| ABLtide | MSSU16 | 25 | 2 | 0.5 | Forward | 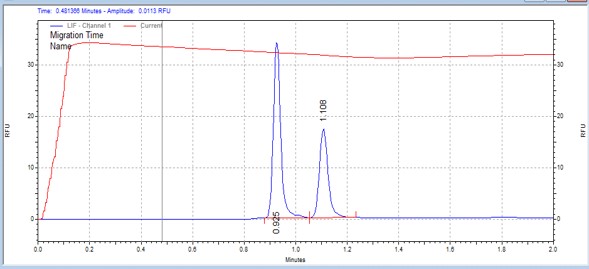 |
| AMARA peptide | MSSU28 | 25 | 2 | 0.5 | Forward | 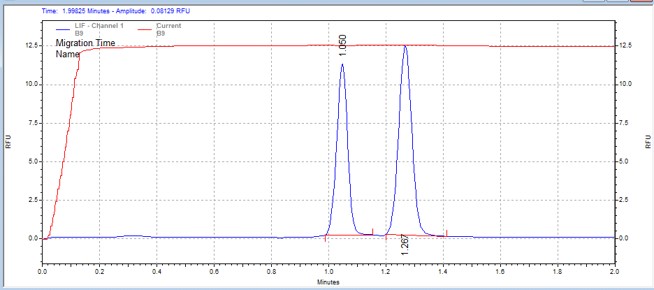 |
| Blk/Lyntide | MSSU04 | 25 | 2 | 0.7 | Forward | 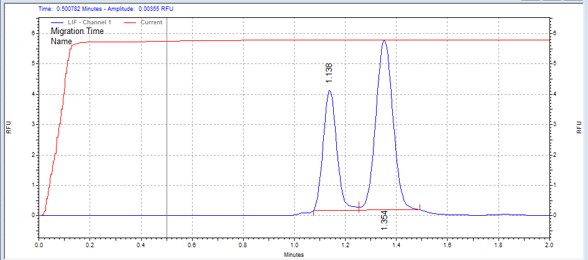 |
| CDC25ctide | MSSU21 | 25 | 2 | 0.8 | Forward | 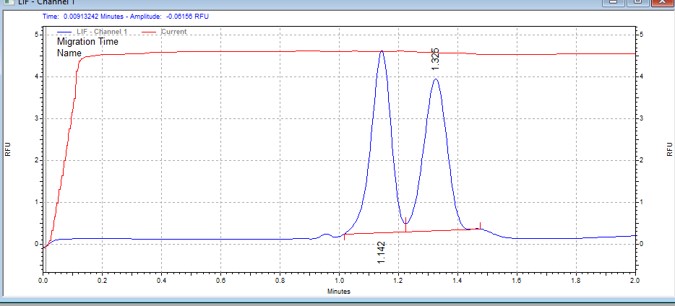 |
| CDK7 peptide | MSSU11 | 25 | 2 | 0.5 | Forward | 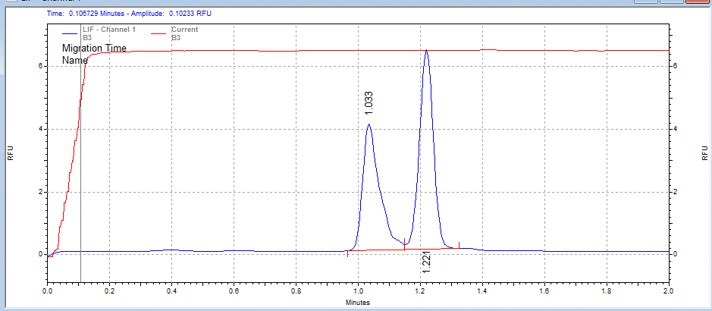 |
| CDK9 substrate | MSSU29 | 25 | 2 | 0.7 | Forward | 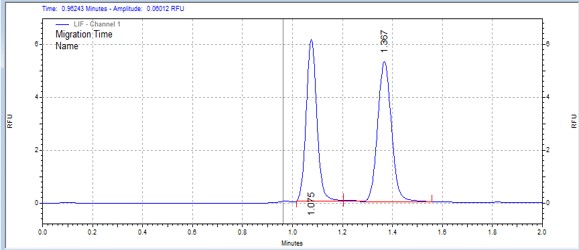 |
| CHKtide | MSSU08 | 25 | 3 | 0.5 | Reverse | 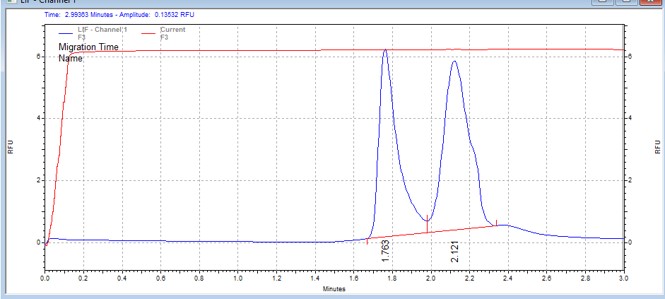 |
| CK2tide | MSSU25 | 25 | 2 | 0.8 | Forward | 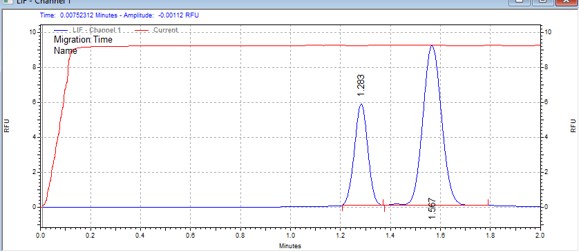 |
| CKtide | MSSU12 | 25 | 2 | 0.5 | Forward | 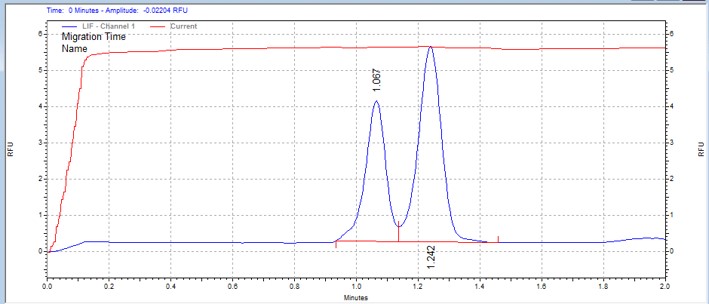 |
| CREBtide-p | MSSU22 | 25 | 2 | 0.5 | Forward | 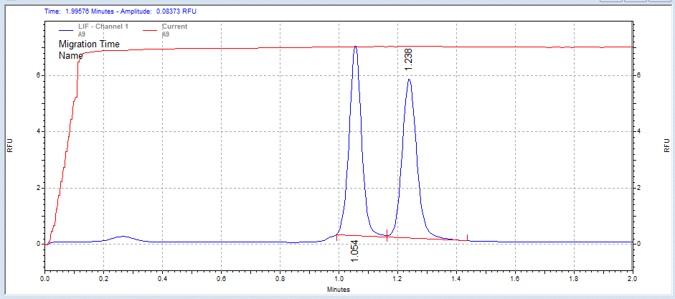 |
| Crosstide | MSSU19 | 25 | 2 | 0.5 | Forward | 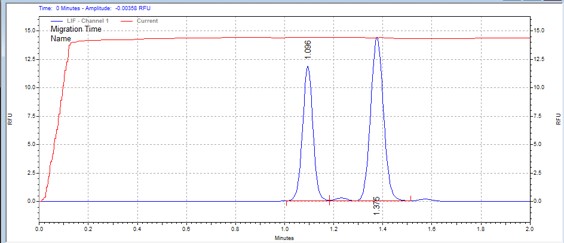 |
| CSKtide | MSSU02 | 25 | 2 | 0.5 | Forward | 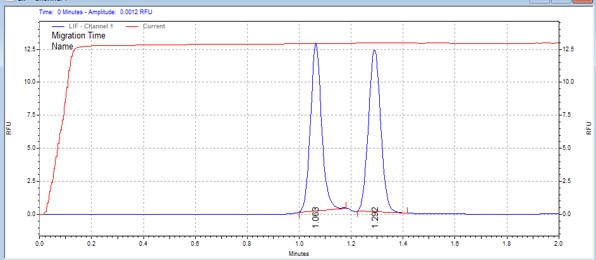 |
| CTD3 peptide | MSSU30 | 25 | 2 | 0.5 | Forward | 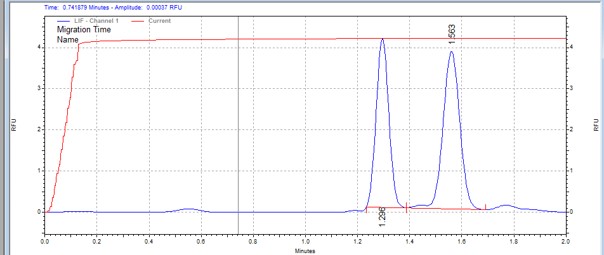 |
| DAPK1tide | MSSU17 | 16 | 3 | 0 | Forward | 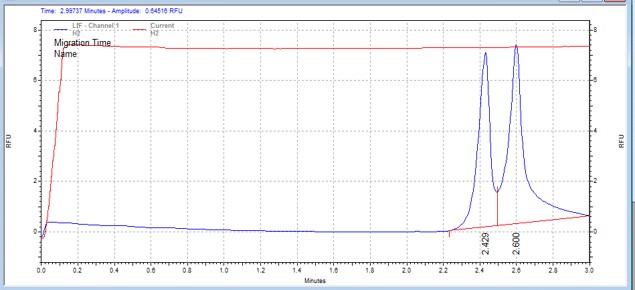 |
| DYRKtide-F | MSSU06 | 25 | 2 | 0.2 | Forward | 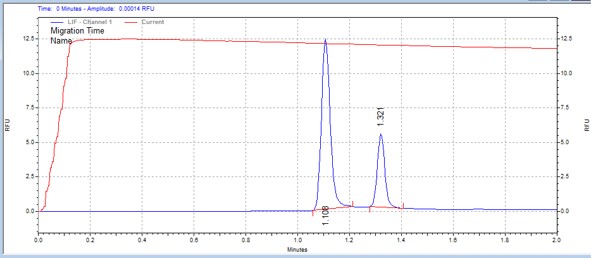 |
| EEF2Ktide | MSSU40 | 25 | 2 | 0.5 | Forward | 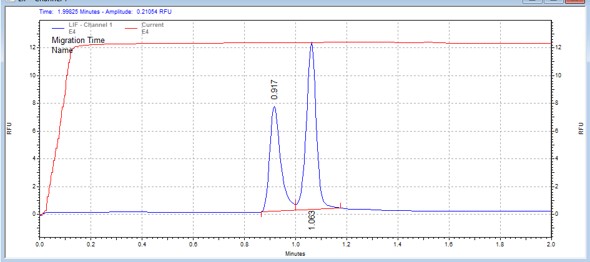 |
| EGFR-derived peptide | MSSU31 | 25 | 2 | 0.8 | Forward | 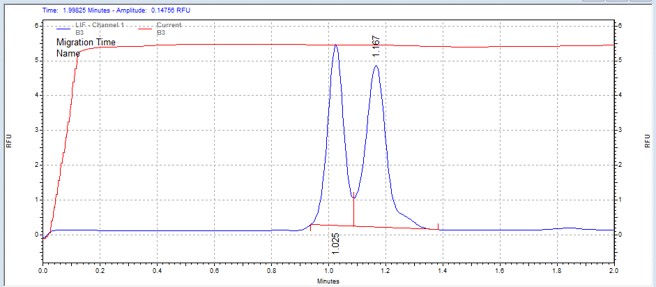 |
| GS peptide | MSSU05 | 25 | 1.5 | 0.8 | Forward | 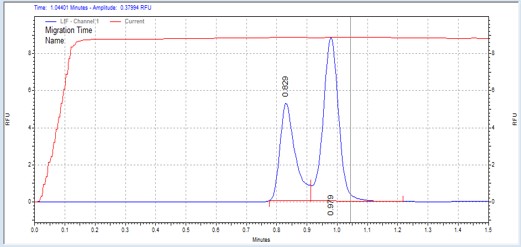 |
| H2A peptide | MSSU47 | 25 | 2 | 0.5 | Forward | 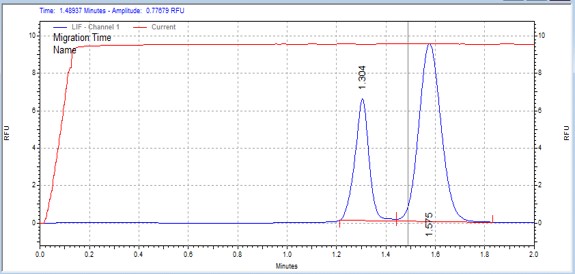 |
| Histone H3 peptide | MSSU23 | 25 | 2 | 0.3 | Forward | 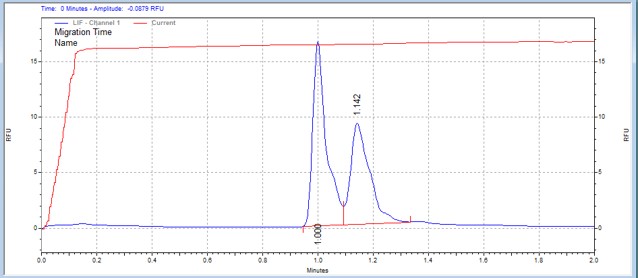 |
| IRAK1 peptide* | MSSU41 | 25 | 1.4 | 0.5 | Forward | 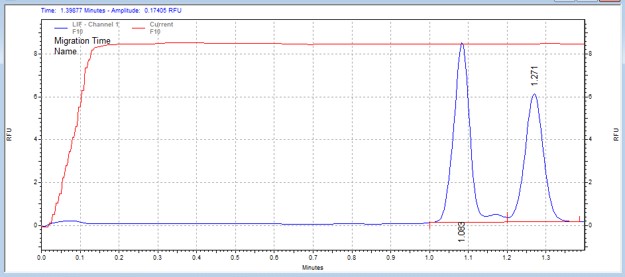 |
| IRS1 | MSSU07 | 25 | 2 | 0.5 | Forward | 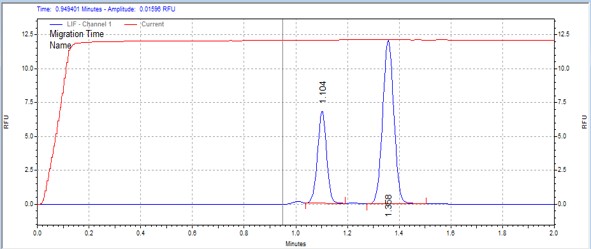 |
| IκBα peptide | MSSU42 | 25 | 2 | 0.8 | Forward | 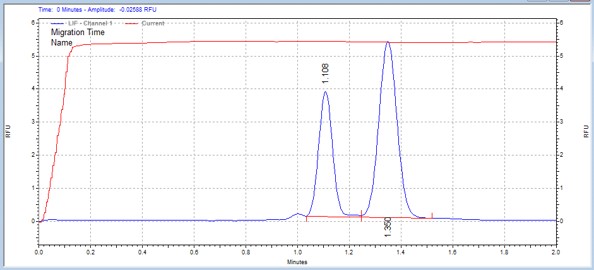 |
| JAK1 substrate peptide | MSSU32 | 25 | 2 | 0.8 | Forward | 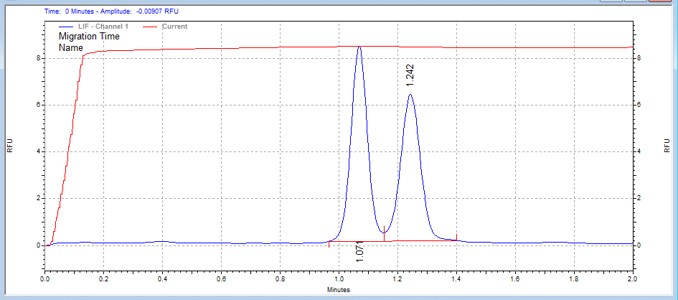 |
| Kemptide | MSSU09 | 25 | 2 | 0.8 | Forward | 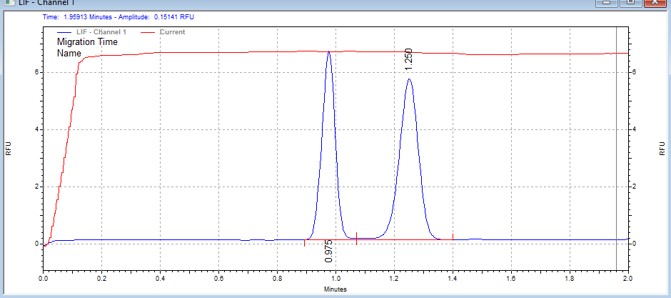 |
| LIMKtide | MSSU24 | 25 | 2 | 0.5 | Forward | 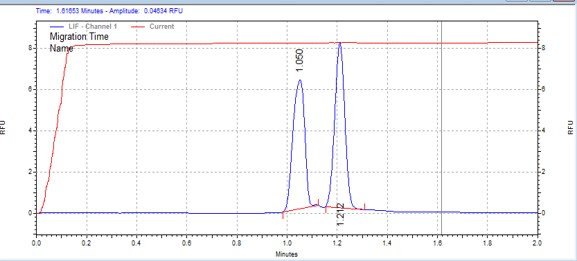 |
| LRRKtide | MSSU48 | 25 | 2 | 0 | Reverse | 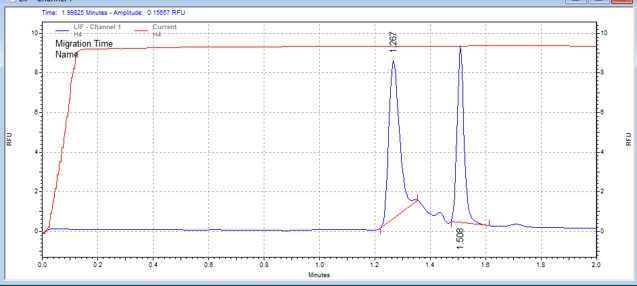 |
| MCM2 peptide | MSSU33 | 25 | 2 | 0.5 | Forward | 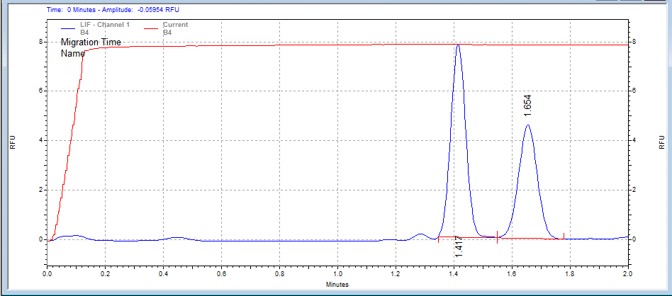 |
| MLCtide | MSSU43 | 25 | 2 | 0.5 | Forward | 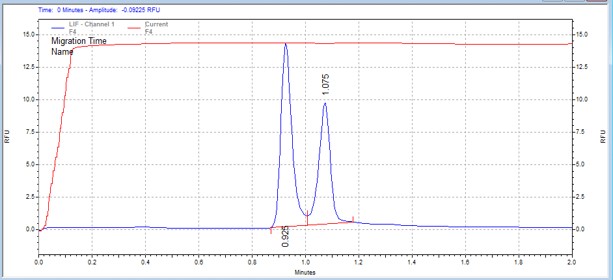 |
| Modified Erktide | MSSU03 | 25 | 2 | 0.5 | Forward | 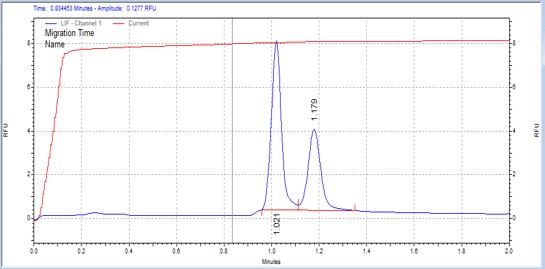 |
| Modified Histone H1 | MSSU13 | 25 | 2 | 0.5 | Forward | 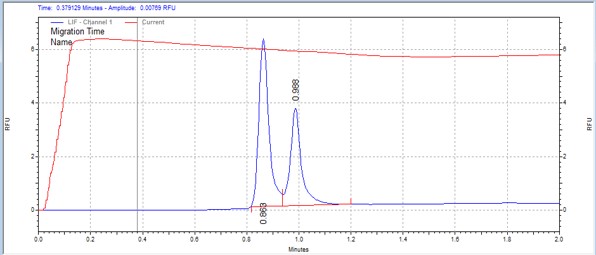 |
| Modified IκBα-derived peptide | MSSU34 | 25 | 2 | 0.8 | Forward | 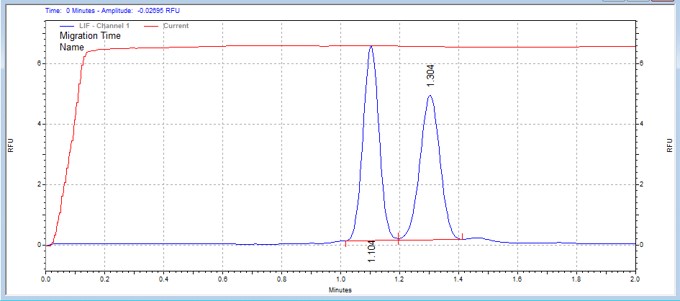 |
| Moesin-derived peptide | MSSU18 | 25 | 2 | 0.8 | Forward | 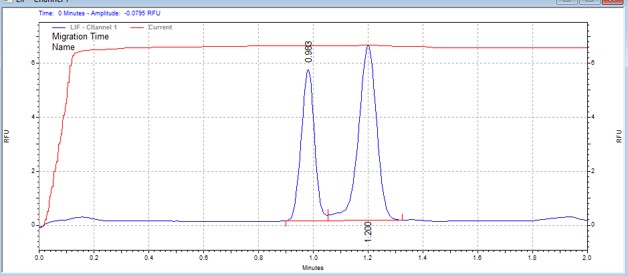 |
| PDHKtide | MSSU35 | 25 | 2 | 0.8 | Forward | 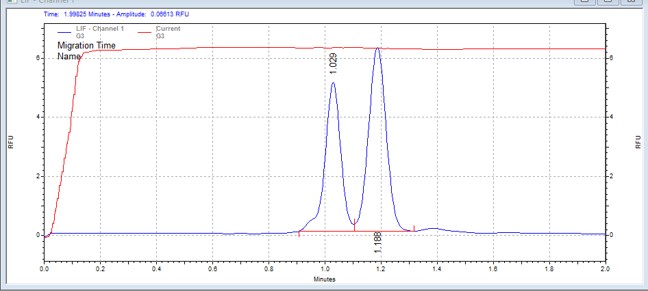 |
| PKC peptide | MSSU10 | 25 | 2 | 0.2 | Forward | 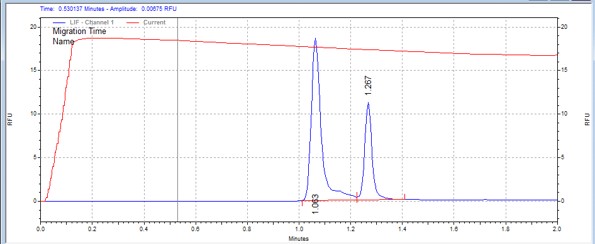 |
| RS peptide | MSSU36 | 25 | 2 | 0.5 | Forward | 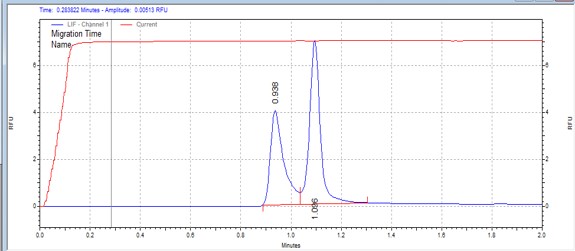 |
| S6K peptide(N-FL) | MSSU20 | 25 | 2 | 0.5 | Forward | 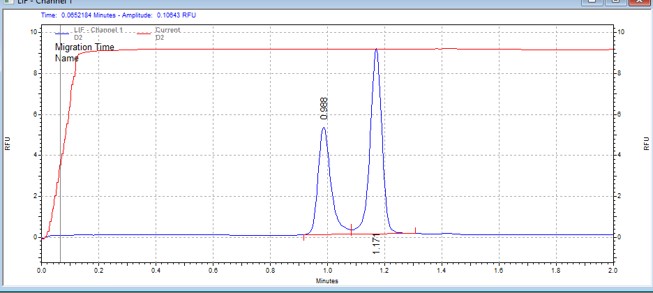 |
| S6K2 peptide | MSSU14 | 25 | 2 | 0.5 | Forward | 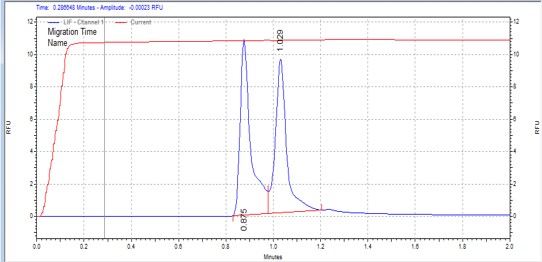 |
| SAMS peptide | MSSU26 | 25 | 2 | 0.5 | Forward | 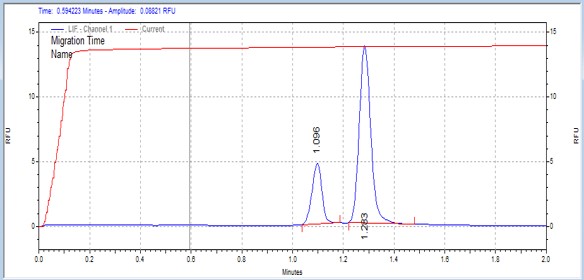 |
| SGKtide | MSSU15 | 25 | 2 | 0.5 | Forward | 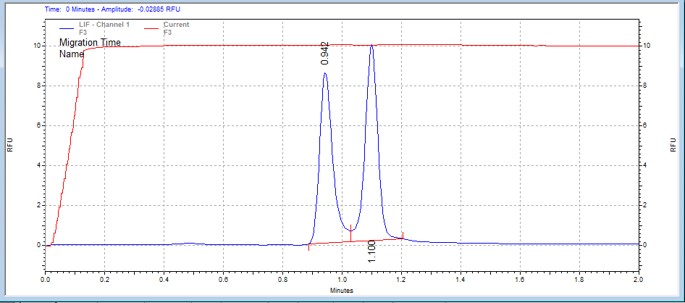 |
| SPAKtide | MSSU27 | 25 | 2 | 0.5 | Forward | 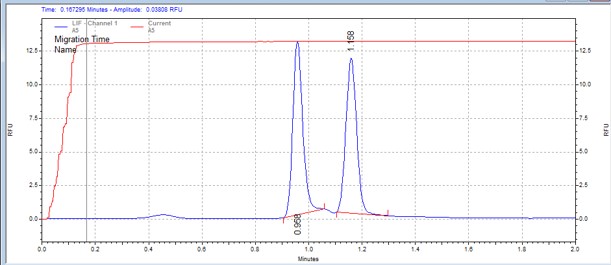 |
| Srctide | MSSU01 | 25 | 2 | 0.5 | Forward | 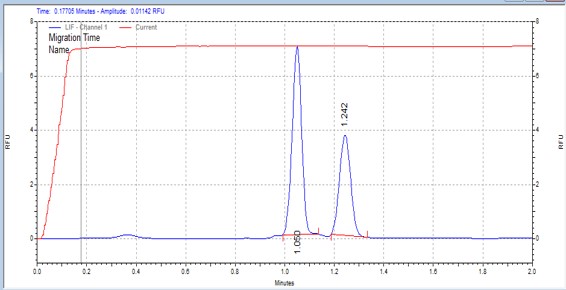 |
| Synapsin peptide | MSSU45 | 25 | 2 | 0.8 | Forward | 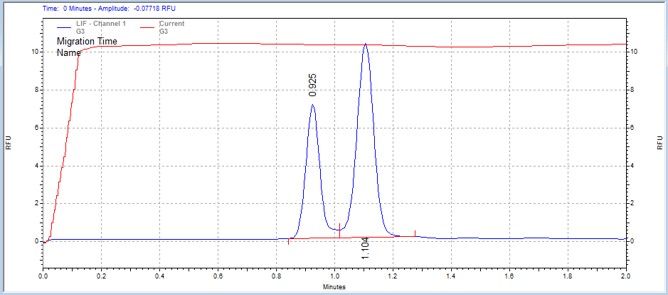 |
| T308tide | MSSU38 | 25 | 2 | 0.5 | Forward | 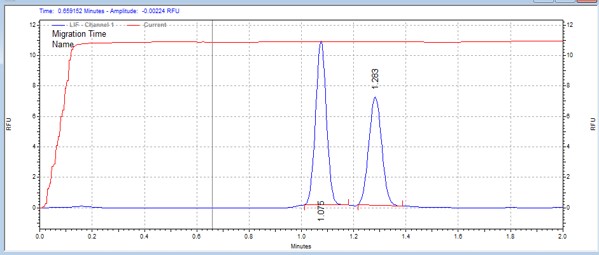 |
| TAOKtide | MSSU46 | 25 | 2 | 0.8 | Forward | 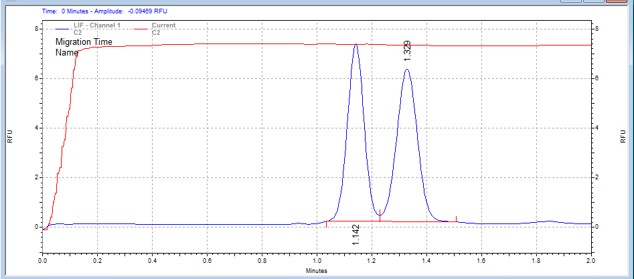 |
| WASP peptide | MSSU39 | 25 | 2 | 0.8 | Forward | 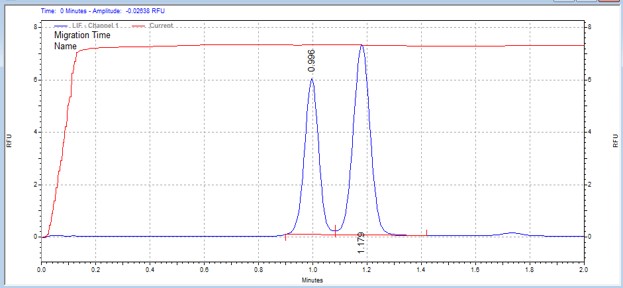 |
Separation Buffer : 100 mM HEPES pH7.5, 10 mM MgCl2, 0.01 % Triton X-100, 10 mM EDTA-2Na, 1 % DMSO
*To remove the peak observed at approximately 1.5 minutes following a single well electrophoresis, an additional step of electrophoresis using Separation Buffer is required.
