- Home
- Cell-Based Assay Services
- NanoBRET™ TE Intracellular Kinase Assay Services
- NanoBRET™ TE Intracellular Kinome-Wide Profiling Service (K192)
NanoBRET™ TE Intracellular Kinome-Wide Profiling Service (K192)
Incorporating a cell-based assay in hit identification often provides advantages over biochemical assays when selecting and characterizing key compounds. As an example, membrane permeability and cytotoxicity, which cannot be determined by biochemical methods using isolated purified proteins, can be assessed in a cellular environment.
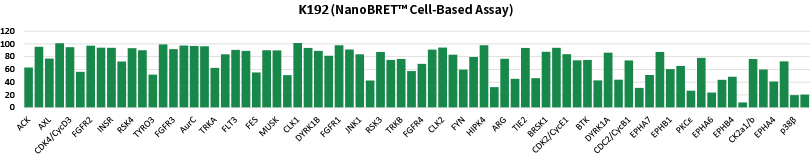
In this cell-based panel profiling service, your compound is evaluated collectively against 192 full-length kinases individually expressed in HEK293 cells under uniform assay conditions using the same tracer. Your compound’s interaction against each target in the cellular environment is quantified, allowing comparison of its efficacy against the primary target(s) and possible off targets.
NanoBRET™ Target Engagement (TE) Intracellular Kinase Assay System (Promega) is used in this broad panel of 192 kinase targets (see list), each expressed intracellularly by transient transfection. Compound evaluation against all targets is performed simultaneously at 1 compound concentration in duplicate, under the same assay conditions. Follow up IC50 determinations also available.
Collectively identify compound properties against 192 kinases, using uniform assay conditions!
Learn what can't be learned in biochemical assays!
Comparison data using 1μM Crizotinib in a biochemical assay (Mobility Shift Assay) and NanoBRET™ K192 cell assay panel (36 selected targets shown)
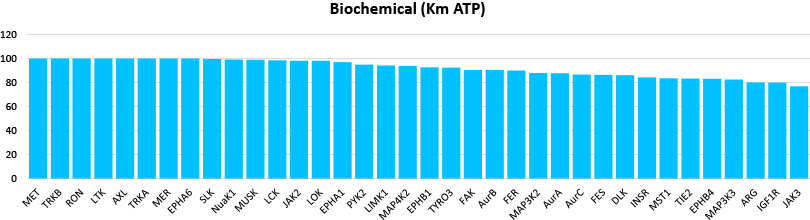
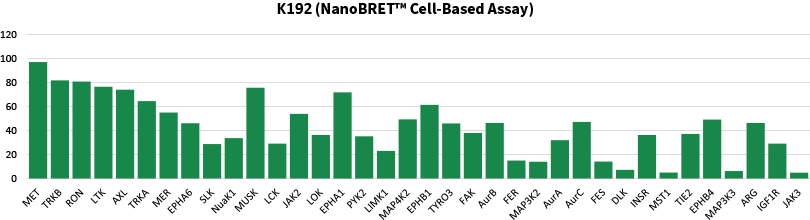
Comparison data using 0.3μM CC1 inhibitor in a biochemical assay (Mobility Shift Assay) and NanoBRET™ K192 cell assay panel
192 Panel Kinase Targets
- <Guide for Service Customers>
- Service Flow
- Compound Handling and Shipping
- Application Form for NanoBRET™ Cell-based Assay
